Auckland Bioengineering Institute
Applications for 2025-2026 open on 1 July 2025
The role of digital health tools and improving Pacific health and wellbeing
Project code: ABI001
Supervisor(s):
Discipline:
Auckland Bioengineering Institute
Project
This project will explore how digital health tools can be used to improve the health and wellbeing of Pacific communities, particularly when using artificial intelligence. There is an opportunity to work on an AI tool to respond to a health topic for Pacific communities.
Ideal student
A student that is knowledgeable with computer science or building software or has developed AI tools would be handy. Otherwise open to med students or 3rd year health science students.
From Researcher to Kairangahau for Tangata Tiriti in Aotearoa
Project code: ABI002
Supervisor(s):
Discipline:
Auckland Bioengineering Institute
Project
The aim of this project is to provide evidence to support our Māori-led approach as a means to influence Kairangahau practice.The research will provide evidence to support our innovative community based model of health research engagement.
Ideal student
Experience working or engaging with Maori-health an advantage. Open to Maori or non-Maori candidates.
Mate hoto hau and hapūtanga Sleep apnoea and pregnancy
Project code: ABI003
Supervisor(s):
Discipline:
Auckland Bioengineering Institute
Project
This project would likely be of greatest interest to a medical student or a student interested in health, specifically wāhine health. Any needed skills can be taught. We would like the student to be able to build whakawhanaungatanga with hapū Māmā and be comfortable discussing sleep, pregnancy, and our study, which we will teach them about.
Information about our study (for potential participants) is available at sleeppregnancy.auckland.ac.nz
Hearing health and research communication
Project code: ABI004
Supervisor(s):
Michelle Pokorny
Discipline:
Auckland Bioengineering Institute
Project
This is a summer studentship project jointly hosted by Dr Michelle Pokorny (Audiology kairangahau, Manukau superclinic) and Dr Haruna Suzuki-Kerr (Ear Physiology kairangahau, UoA).
Michelle is working on multiple projects using AI and other emerging tools to efficiently diagnose and treat hearing loss, while Haruna is working on developing a new diagnostic and treatment medical device for hearing loss.
Michelle and Haruna are both passionate about hearing health and sharing their research with the community, however, in recent years they have noticed a need for more open, community focused resources.
The role
The aim of this summer studentship is to produce appropriate information material to communicate research and health information accurately and clearly that can be used in hearing screening, and research showcases with the community. Over the summer, the summer studentship scholar will work with Michelle and Haruna to learn about hearing health, research and clinical activities to produce these materials.
Exploring Natural Product Testing for Health Tech Applications
Project code: ABI005
Supervisor(s):
Discipline:
Auckland Bioengineering Institute
Project
This project offers a unique opportunity to explore how natural products can be tested using modern biological assays.
The role
The student will gain hands-on experience working with human and mouse cell lines to investigate anti-inflammatory effects, barrier function, and toxicity.
Alongside lab work, there will be scope to explore the commercial and health tech potential of these assays — including identifying market needs and opportunities for future service development.
Ideal student
This is a great fit for students interested in combining lab science with real-world impact.
Unlocking Temporal EEG Signatures of Neonatal Hypoxic-Ischemic Brain Injury with Machine Learning
Project code: ABI006
Supervisor(s):
Associate Professor Joanne Davidson
Discipline:
Auckland Bioengineering Institute

Project
Newborns, particularly those born pre-term, face a substantial risk of brain injury after hypoxia-ischaemia (HI), a period of reduced oxygenation and cerebral blood flow that can lead to lifelong neurodevelopmental disability. Although neonatal care has advanced, dedicated neuroprotective and neuro-reparative therapies remain scarce.
HI may arise during labour or in utero, and the resulting injury progresses through distinct pathophysiological phases that span days to weeks. Therapeutic success, as illustrated by hypothermia treatment for term infants, depends on precise timing; our group has shown that the optimal window for cooling lies within the first 6–7 hours after HI, with maximal benefit when initiated in the initial 3 hours of life.
Accurate treatment timing requires biomarkers that reveal both the risk of injury and its evolving stage. Continuous electroencephalography (EEG) provides a bedside measure of cerebral function, and our previous work confirms its utility in tracking HI progression.
The role
Building on this foundation, the summer project will analyse high-resolution EEG recordings from pre-term and full-term fetal sheep to characterise how spectral features change after HI and under different therapeutic interventions.
Aims
The project will address three objectives:
- Quantify the temporal evolution of EEG spectral power and connectivity during each injury phase.
- Compare EEG biomarkers between pre-term and full-term models to identify gestational age–specific patterns.
- Assess how candidate interventions modulate these EEG signatures and refine the timing of therapy.The insights gained will guide future translation of EEG-based biomarkers to neonatal intensive care and support the development of phase-specific treatments for vulnerable infants.
Timing
4 months
Desired skills
We seek a motivated student with a background in engineering, computer science, or biomedical engineering, with a strong interest in medical data analysis and computational neuroscience, as well as proficiency in Python or Matlab. Experience in machine/deep learning, and/or medical signal/image analysis is highly beneficial.
Investigating Epicardial Adipose Tissue Geometry and Shape: Unraveling Its Role on Atrial Fibrillation
Project code: ABI007
Supervisor(s):
Fan Feng
(Supporting: Yun Gu)
Discipline:
Auckland Bioengineering Institute
Project
Atrial Fibrillation (AF) is the most prevalent cardiac arrhythmia, leading to increased risks of stroke, heart failure, and other cardiovascular complications. Emerging evidence suggests that the geometry and shape of Epicardial Adipose Tissue (EAT) play a critical role in the development and persistence of AF. However, the exact mechanisms through which EAT geometry influences atrial function and AF onset remain poorly understood, limiting the effectiveness of current therapeutic approaches.
EAT is a unique fat depot located between the myocardium and the visceral pericardium. Its proximity to the heart's atrial chambers suggests a significant influence on atrial electrophysiology and structural integrity. Variations in the geometry and shape of EAT may contribute to atrial remodeling, a key factor in AF pathogenesis.
The aims of this study
- Utilize Deep Learning to Segment Epicardial Adipose Tissue (EAT) and the Four Cardiac Chambers from Large-Scale CT Data
- Establish Statistical Shape Models to Study the Geometry Distribution of Epicardial Adipose Tissue
- Conduct clinical studies to assess how variations in EAT geometry influence ablation outcomes and develop personalized ablation approaches based on patient-specific EAT characteristics
Desired skills
Ideal candidates will have solid background in Engineering/Mathematics and programming skills. Experience in medical imaging will be beneficial.
Machine Learning for Enhanced Glial Cell Detection in Hypoxic–Ischaemic Brain Injury Images
Project code: ABI008
Supervisor(s):
Associate Professor Joanne Davidson
Discipline:
Auckland Bioengineering Institute
Project
Insufficient oxygen and cerebral blood flow around birth can precipitate hypoxic-ischemic encephalopathy (HIE), triggering a complex inflammatory cascade in which microglia and astrocytes play central roles.
The role
This summer project will accelerate computational investigations of these glial cells by assembling a meticulously annotated image repository and creating augmentation pipelines that broaden its research value. You will assist in the annotation process using a mix of manual labeling and semi-automated techniques, and potentially also working on extensions for the segmentation GUI that will benefit the broader community.
Following dataset curation, you will explore and implement novel data augmentation strategies aimed at expanding the dataset without the use of manual annotations. These augmentations will be validated using basic machine learning models (e.g., CNN or transformer based segmentation models).
By the conclusion of the project, you will have developed robust methodologies and user-friendly tools that substantially accelerate glial-cell investigations in neonatal brain-injury research.
Required skills
- Proficient Python programming
- Familiarity with image processing libraries (e.g., OpenCV, scikit-image)
- Exposure to the PyTorch machine learning frameworks (desirable, but not required)
- Attention to detail
- Teamwork
- A basic knowledge of biology, including microglia and astrocytes
Integrated modular cardiovascular system gadgets for physiology exploration
Project code: ABI009
Supervisor(s):
Dr Beatrice Ghitti
Dr Finbar Argus
Discipline:
Auckland Bioengineering Institute
Project
The cardiovascular system is a complex and dynamic system whose behavior varies significantly across individuals. Computer models and simulations provide powerful tools to study the circulatory system and improve understanding of its function and response to physiological changes, offering a platform to visualize and analyze cardiovascular dynamics in a controlled and interactive way.
The role
This project aims to design and build interactive cardio-vascular modules using Raspberry Pi devices as an educational and prototyping tool. Each Raspberry Pi will represent a component of the cardiovascular system (e.g., a blood vessel or the heart) and run embedded simulations. These devices will be interconnected physically and digitally to replicate physiologically relevant, user-defined topologies of portions of the cardiovascular system. Specialised communication protocols will be implemented for real-time data exchange between devices. Each unit will solve a computer model of blood flow, allowing simulation of pressure and flow waveforms propagating through the system. The platform will also integrate real-time physiological input (e.g., heart rate, PPG signals) acquired from wearable sensors, to explore subject-specific responses.
Outcome
This project will result in a scalable, hands-on educational tool that enables users to construct, modify, and interrogate cardio-vascular systems – integrating physiology, modeling, and hardware-level programming.
Requirements
We are looking for highly motivated students with a background in engineering, computer science, mathematical modelling, and/or physics. Prior programming experience (ideally C++/Python) is preferred.
Skills gained
Sensor interfacing and integration with computational models; Efficient model simulation schemes; Hemodynamic modelling.
Verification of paediatric clinical gait workflow
Project code: ABI010
Supervisor(s):
Julie Choisne
Discipline:
Auckland Bioengineering Institute
Project
Skeletal growth in children involves complex interactions between biological and mechanical factors. The majority of growth occurs within the first few years, with continued growth until fusion of growth plates, which can be up to 20 years of age in the lower limbs.
Typically developing children experience programmed changes in long bone dimensions as they grow, while atypical development, for example, in children with neuromuscular diseases, can lead to altered skeletal growth, affecting hip stability, gait, and limb alignment. Because atypical development can be very heterogeneous depending on the initial diagnosis, computational modelling is often used to inform treatment. However, computational models of children are quite limited and most of the current models are based on adult anatomy.
The role
We have developed an automatic workflow for performing gait analysis in a paediatric population. This workflow uses shape modelling and musculoskeletal modelling to calculate the kinematics and kinetics of gait.
This project will validate the workflow using an already existing dataset of 50 children with gait pathologies. The dataset includes motion capture and partial medical imaging which will be used to predict musculoskeletal models using this workflow. The output kinematics and kinetics of gait will then be compared between three methods of model generation: linear scaling, prediction from motion capture markers, and prediction from partial medical imaging data.
Required skills
Python
Beating heart disease – non-contact imaging for cardiovascular disease – blood pressure pilot study
Project code: ABI011
Supervisor(s):
Alex Dixon
Poul Nielsen
Supporting: Andrew Taberner, Martyn Nash
Discipline:
Auckland Bioengineering Institute
Project
Cardiovascular disease (CVD) affects millions worldwide and is the leading cause of mortality. Devices that can efficiently and non-invasively provide early and clinically useful diagnostic information may improve patient quality of life and help reduce CVD morbidity.
Our group has created a camera-based imaging system that estimates the carotid artery and jugular venous pressure waveform by measuring the deformation of the skin on the neck due to the vessel pulsations. The results to date highlight the potential for this system to be used as a non-invasive diagnostic tool for cardiovascular disease.
The role
This summer studentship project will focus on key developments for the current system. The project will be based on evaluating the techniques with mobile high-speed video recording capabilities, including spatial video and stereoscopic imaging. These investigations will be conducted alongside quantitative extraction of blood pressure-related biomarkers. Overall, these advancements will enable the translation of this research toward a home-based healthcare tool.
Project aims
The main objectives of this project will be to:
- Implement and acquire videos from a mobile phone
- Perform experiments for video and blood pressure acquisition
- Analyse 3D video/deformation and identify BP imaging biomarkers
- Analyse and present data from experiments.
Ideal student
The project will suit students interested in translational research and will involve hardware development with cameras and optics, bioinstrumentation, imaging experiments, and signal processing/data analysis. Prior medical imaging experience is not required.
Outcome
This project will enable the development of more efficient and effective strategies for the diagnosis and monitoring of cardiovascular diseases for patients in the clinical setting and at home.
Waxing and Waning of Intestinal Slow Wave Activity
Project code: ABI012
Supervisor(s):
Nipuni Nagahawatte
Recep Avci
Discipline:
Auckland Bioengineering Institute
Project
Your gut contracts to enable the breaking down, mixing, and propulsion of food within your gut. These movements or contractions are coordinated by rhythmic electrical events known as slow waves. It is hypothesised that waxing and waning is related to competing pacemakers operating in and out of phase. High-resolution GI slow wave mapping allows the mapping of spatial propagation patterns of small intestine slow wave activity.
The opportunity now exists to:
- Investigate the spatial distribution of waxing and waning activity in the small intestine, to determine if it is a spatial effect of competing pacemakers.
- Compare the prevalence and morphology of waxing and waning in different regions of the small intestine.
- Determine the frequency content of in vivo waxing and waning recordings, combined with the spatial distribution, to investigate potential of coupling of multiple dominant frequencies.
Ideal student
The student should ideally have a strong interest and background in signal processing and data analysis. Proficiency in MATLAB is preferred. No previous experience with experimental physiology is required.
High-performance, low-cost stereo reconstruction
Project code: ABI013
Supervisor(s):
Supportong: Alex Dixon, Robin Laven
Discipline:
Auckland Bioengineering Institute
Project
Stereo reconstruction uses images obtained from different views to infer the 3D shape of an object. Many high-resolution images are required for accurate reconstruction. If the object is rapidly changing shape (e.g. dynamic facial expressions), high-frame-rate cameras are required. Many high resolution, high frame rate cameras are typically costly and bulky. However, small low-cost cameras are becoming available to interface with small, powerful computers such as Raspberry Pi and NVIDIA Jetson.
The role
This project will explore suitable camera and computer combinations to build a relatively inexpensive but high-performance multi-camera stereo imaging device. Images from the device will be used to calibrate the system, using our state-of-the-art software, enabling very high-resolution reconstructions of dynamically changing shapes.
Design and development of an endoscopic training tool
Project code: ABI014
Supervisor(s):
Leo Cheng
Peng Du
Discipline:
Auckland Bioengineering Institute
Project
Endoscopy is a minimally invasive procedure used to visualise the interior of the gastrointestinal tract using a flexible tube with a camera. It aids in diagnosing conditions such as tumours and ulcers. Therapeutic interventions, such as polyp removal or bleeding control, can also be performed endoscopically. Endoscopic bariatric procedures, such as endoscopic sleeve gastroplasty and intragastric balloon placement, promote weight loss without external incisions.
Endoscopy training typically involves a combination of physical and virtual reality simulation-based practice, supervised procedures on patients, and structured feedback to develop technical and diagnostic skills. Modern methods may include virtual reality simulators and competency-based assessments to enhance learning and ensure proficiency.
The role
This project will design and create a custom endoscopic training tool. It will include two main geometries: an idealised cube domain and a realistic stomach shaped domain.
Possible training tasks may include, grasping, manipulating and moving objects between different regions of the domain.
Ideal student
The student should ideally have a strong interest and background in device design, CAD, 3D printing etc. Proficiency in SolidWorks is preferred.
Virtual Guts: Digital Twins for Disorders of Gut-Brain Interaction
Project code: ABI015
Supervisor(s):
Weiwei Ai
Supported by David Nickerson
Discipline:
Auckland Bioengineering Institute
Project
Around one-in-three people live with a disorder of gut-brain interaction (DGBI), such as irritable bowel syndrome, yet our understanding of these conditions remains limited. DGBI can significantly impact quality of life without presenting with any clear disease markers, making diagnosis difficult and time-consuming, often taking years. Even following diagnosis, treatment typically involves a frustrating period of trial-and-error.
The role
But what if clinicians could screen treatments on a virtual version of a patient before prescribing them? This summer research project will help in the development of a digital twin of the gastrointestinal (GI) tract – a cutting-edge computational model that simulates patient-specific GI function.
You’ll contribute to this ambitious goal by building on existing frameworks and applying parameter estimation techniques, a core component of many machine learning applications.
Ideal student
Experience with Python is recommended but not essential. Curiosity and passion for solving real-world problems are a must. If you’re excited about using computational models to transform patient care and want to help tackle a modern health mystery, this project is for you.
Advanced analysis of lymphatic drainage pathways in head and neck melanoma
Project code: ABI016
Supervisor(s):
Supported by Tharanga Don
Discipline:
Auckland Bioengineering Institute
Project
Melanoma is the most aggressive form of skin cancer, known to spread from the skin to nearby lymph nodes, which can lead to to secondary tumours. Understanding this metastatic pattern is crucial for improving treatment planning and patient outcomes.
The role
In this project, students will analyse a large-scale lymphoscintigraphy (LSG) imaging dataset from over 17,000 melanoma patients, focused specifically on the head and neck region. These imaging data have been spatially mapped onto a 3D computational model of the skin and lymph nodes. Additionally, more than 6,000 patients are linked to rich clinical metadata - including demographics, pathology, and long-term follow-up - the Melanoma Institute of Australia, the world’s most comprehensive melanoma database.
Students will apply image analysis and visualisation techniques to uncover patterns in lymphatic spread, with the goal of understanding potential metastatic pathways in head and neck cancer.
Skills desired
Computer programming and statistical methods
Cracking the Gut’s Electric Code
Project code: ABI017
Supervisor(s):
Leo Cheng
Nipuni Nagahawatte
Supported by GI Group
Discipline:
Auckland Bioengineering Institute
Project
Rhythmic electrical signals that govern stomach function are often disrupted in patients with conditions such as nausea and indigestion. Despite their clinical relevance, non-invasive methods for detecting and analysing these signals remain underdeveloped due to weak signal characteristics and the complexity of stomach anatomy. Our research aims to bridge this gap by leveraging cutting-edge tools to map and model the intricate relationship between gastric electrical signals, bioelectromagnetic fields outside the body, and subject-specific anatomy.
Our vision is to revolutionise the diagnosis of gastric disorders by developing reliable, non-invasive tools that clinicians can use to detect and monitor bioelectromagnetic patterns indicative of these conditions.
The role
As a summer student, you will contribute to the computational aspects of this vision, gaining expertise in advanced electrophysiology, bioengineering, and healthcare innovation.
This project aims to develop a mathematical model of gastric electrical signals by optimising source configurations and forward computations. Specifically, you will:
i) Investigate different source configurations and compare their corresponding bioelectromagnetic fields outside the body
ii) Evaluate and compare software packages for solving the forward problem of gastric electrical signals.
Desired skills
Interest in electrophysiological modelling, working knowledge of MATLAB and/or Python, and familiarity with finite element method techniques.
Designing clinical reports for personalised lung disease management
Project code: ABI018
Supervisor(s):
Behdad Shaarbaf Ebrahimi
Supported by Megan Soo, Merryn Tawhai
Discipline:
Auckland Bioengineering Institute
Project
The Auckland Bioengineering Institute’s Lungs and Respiratory System group has developed several workflows that utilise medical images (e.g. CT or MRI) to create personalised computational physiology models of the lungs. These workflows have been used to explore different respiratory disease processes and can become novel clinical tools for monitoring respiratory diseases. To help further prepare our workflows for a clinical hospital setting, we need an automated process for creating personalised clinical reports for each patient.
The role
In this project, we are looking for a student to develop an automated pipeline that will create a summary clinical report that collates different computational modelling results, clinical measurements and medical images for a range of patients with lung diseases.
Skills gained
In this project you will have the opportunity to develop skills in medical imaging analysis, Python coding, and clinical translation.
Actuated printing of conducting polymer sensors
Project code: ABI019
Supervisor(s):
Jadranka Travas-Sejdic
Discipline:
Auckland Bioengineering Institute
Project
Medical devices for diagnosis and rehabilitation are involving ever increasing number of sensors.
The role
In this project we will develop and refine manufacturing capabilities of 3D extrusion printing of conducting polymer sensors for multi-electrode bio-signal acquisitions. However, one of the challenges with a multi-electrode setup is the robustness of the tracks as they are printed onto the substrate material.
To further improve the mechanical robustness of the sensor array, we will develop an approach with incorporating the single-pair twining technique of te raranga me te whatu (traditional weaving and pattern) to produce durable conducting tracks.
Tasks:
1) Identify the key specifications of non-invasive medical electrode applications (e.g. for cardiac, gastric and muscle recordings)
2) Visit automated dispenser setup and learn the extrusion manufacturing technique and its key production parameters
Ideal student
The ideal candidate should be eager to engage and have an interest in engineering and chemistry.
User Study and Data Support for a Digital Human Mental Health Tool
Project code: ABI020
Supervisor(s):
Supported by Kunal Gupta
Discipline:
Auckland Bioengineering Institute

Project
The Tōku Hoa project at the Empathic Computing Lab explores how digital mental health companions can support emotional wellbeing in young people. Built in collaboration with Soul Machines, the Tōku Hoa mobile app features a conversational AI agent that provides personalized support using physiological and contextual inputs from the user’s smartwatch and smartphone. The app enables reflective journaling, emotional check-ins, lightweight goal-setting, and just-in-time support conversations that adapt to the user’s stress or emotional state.
The role
This summer project will support the real-world evaluation study of the Tōku Hoa app, where participants will use the app daily over a four-week period, while physiological and behavioural data are collected passively through the smartwatch and smartphone sensors. The study aims to assess usability, engagement, and emotional expressiveness in naturalistic, everyday settings.
Key Responsibilities
You will contribute to both study operations and data management tasks, including:
- Participant Support and Technical Setup
- Assist in onboarding participants and setting up the smartwatch-app system
- Troubleshoot basic technical issues (e.g., device sync, connectivity)
- Maintain participant tracking and check-in logs
- Data Preprocessing and Management
- Clean and organize multimodal data from app usage and sensor logs (e.g., HRV, activity, journaling entries)
- Tag and anonymize datasets using consistent conventions
- Documentation and Reporting
- Maintain clear logs of data processing steps and participant study sessions
- Contribute to internal summaries on app engagement and study adherence
Deliverables
- Cleaned and anonymized dataset from smartwatch and app logs
- Internal report or slide deck summarizing key usage metrics
- Contribution to usability issue log or app improvement suggestions
Learning Outcomes
- Real-world experience supporting a human-cantered technology evaluation
- Practical skills in handling wearable and app-based data
Exposure to applied affective computing and personalized mental health support - Understanding of user-cantered research methods in digital wellbeing
Ideal Candidate
- Basic experience with data analysis tools (e.g., Python/Pandas or R)
Reliable and detail-oriented - Interest in mental health, UX, or AI-driven digital interventions
- Bonus: Familiarity with Android systems, physiological data, or HCI research
Eye motion analysis by machine learning
Project code: ABI021
Supervisor(s):
Supported by Dr Jason Turuwhenua
Discipline:
Auckland Bioengineering Institute
Project
You will neeed to be familiar with Python programming and computer vision, AI, Deep learning and GenAI – full time internship.
Evaluating Full Body Kinematics with and without Head-Mounted AR/VR Display
Project code: ABI022
Supervisor(s):
Supported by Ted Yeung
Discipline:
Auckland Bioengineering Institute
Project
Walking, or gait, is more mentally demanding than many people realise—it’s not just an automatic process. Performing another task while walking, such as using a phone or wearing a head-mounted display (HMD), can interfere with memory and cause people to move differently in compensation. This can affect their awareness of their surroundings and the amount of mental effort they’re using.
The role
This project will use a rich dataset to investigate whether wearable sensors could be used in aiding the detection of the increased mental effort. You will aid the research team in processing the optical and inertial mocap data and helping evaluate the machine learning model that estimates whether the person’s movement is normal.
Skills Required
Python, multiprocessing, machine learning
Make computational biology faster through automatic mathematical conversion
Project code: ABI023
Supervisor(s):
Alan Garny
Discipline:
Auckland Bioengineering Institute
Project
Computational biology requires significant expertise to implement. We have developed software to make this process easier, but support for models with Differential Algebraic Equations (DAE) could be improved by having them automatically converted to Ordinary Differential Equations (ODE), whenever possible.
The aim:
- Identify some types of DAE models that can be converted to an ODE model
- Develop an algorithm for such a conversion or conversions using the abstract syntax tree (AST) representation of a model's equations
- Implement that algorithm in C++ in libCellML (alternatively, the student can implement a proof of concept in Python using libCellML's Python bindings)
- Use libCellML to generate some C (or Python) code for both the original (DAE) model and the converted (ODE) model, to confirm that both versions of the model yield the same results
Ideal student
This project is aimed at a student with a strong background in both calculus and software development (be it in C++ and/or in Python).
Extra info about the software
CellML (https://cellml.org/) is a standard used to encode pure algebraic models, ODE models, and DAE models. libCellML (https://libcellml.org/) is a C++ library (with Python bindings) for tool developers who want to leverage CellML in their applications.
libCellML comes with an analyser which job is to determine the type of a model, i.e. whether it is a pure algebraic model, an ODE model, a DAE model, or an invalid model. DAE models are the most costly to compute. However, through mathematical manipulation, some DAE models can be converted to an ODE model, making the resulting model significantly faster to compute.
Building a pioneering child health study with community participation
Project code: ABI024
Supervisor(s):
Vickie Shim
Supported by Maryam Tayebi and Samantha Holdsworth
Discipline:
Auckland Bioengineering Institute
Project
Paediatric imaging studies help understand human anatomy as we grow and develop. They can inform our understanding of normal anatomy and the epidemiology of disease, and help us identify early biomarkers of diseases.
The University of Auckland and Mātai Medical Research Institute are developing an ambitious, long term study to follow up a cohort of children in Tairāwhiti district. Advances in MRI technology make this an opportunity to understand the human body in greater detail than before.
This study is being developed with community and academic collaborators. Kaupapa Māori values will be central to the full study.
The role
The research student will assist in designing the full study, building relationships with community stakeholders in the district along with academic collaboration.
Ideal student
We are looking for a student who is interested in child health and Māori health, community participation, and research design. Being a strong communicator will be essential.
The research will be conducted in Tairāwhiti, so the student should be able to spend majority of the internship time in Gisborne.
Skills gained
- Understanding research processes
- Working with community stakeholders
- Academic writing
Evaluating Targeted MRI for Enhanced Sensitivity and Specificity
Project code: ABI025
Supervisor(s):
Vickie Shim
Supported by Maryam Tayebi, Samantha Holdsworth, Dan Cornfeld, Paul Condron, Gil Newburn, Graeme Bydder, and Miriam Scadeng
Discipline:
Auckland Bioengineering Institute
Project
This summer research project will focus on analysing images from a novel Ultra-High Contrast (UHC-)MRI method developed by our team, to detect and visualise neuroinflammation in the brain. This cutting-edge technique aims to enhance the specificity and sensitivity of MRI in identifying neuroinflammatory processes associated with various neurological conditions such as multiple sclerosis, Alzheimer's disease, and traumatic brain injury.
The role
The project will involve comparing UHC-MRI images acquired on a clinical cohort of participants and comparing their efficacy with standard clinical MRI. The overall goal of the research team is to establish a reliable, non-invasive imaging biomarker for neuroinflammation that can improve diagnosis, monitor disease progression, and evaluate the efficacy of therapeutic interventions.
As the research will be mainly based in Tairāwhiti, the student is required to spend most of the internship in Gisborne.
Skills recommended
MATLAB, image processing, data analysis
Co-designing an interactive digital storybook for advancing breast health awareness
Project code: ABI026
Supervisor(s):
Supported by ABI Breast Biomechanics Research Group
Discipline:
Auckland Bioengineering Institute
Project
Breast cancer affects 1 in 9 women in Aotearoa New Zealand. Raising awareness of breast cancer issues and educating all women about breast health is vitally important and can contribute to early detection and outcomes.
The role
The overall goal of this project is to co-design an interactive digital storybook with the community to communicate key concepts for advancing breast health awareness. This information resource will integrate visualisations from breast cancer research to help communicate information to a range of audiences including the general public, patients, and whanau.
You will join a multi-disciplinary team of community engagement specialists, web application developers, bioengineers, clinicians, and nurse educators from the New Zealand Breast Cancer Foundation.
A Dynamic Heartbeat Editor
Project code: ABI027
Supervisor(s):
Weiwei Ai
Supported by Ben Allen (PhD candidate)
Discipline:
Auckland Bioengineering Institute
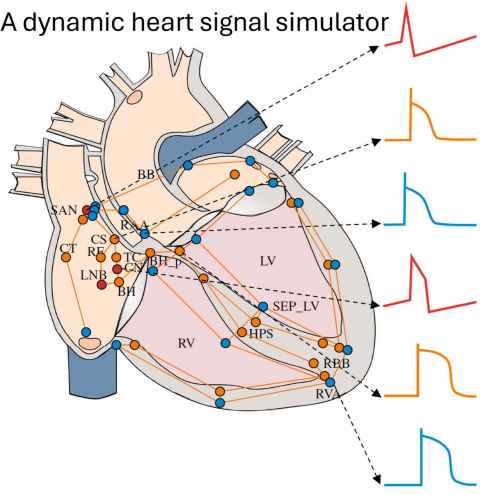
Project
A real-time heart signal simulator has been developed that can interact with medical devices. This is generating interest in the medical device industry, and it is part of a collaboration with MathWorks Inc (the software company that produces Matlab and Simulink).
The role
This Summer Research project will develop an editor to dynamically alter features of the computer models driving the heart simulator and display the effects of these changes on whole heart function. For example, electrical conduction is altered in parts of the model to mimic radio-frequency- or cryo-ablation, so that the impact of ablation therapy on electrical sequences in the heart can be observed and evaluated.
Skills required
1) An interest in software and developing applications and interfaces
2) An eye for design and visual appeal
3) Some knowledge of algorithm design and implementation for efficiency
4) Experience in Matlab and perhaps Simulink
Periscope3D - A Robotic 3D Sensing System for Enhanced Remote Physical Collaboration
Project code: ABI028
Supervisor(s):
Supported by Mark Billinghurst
Discipline:
Auckland Bioengineering Institute

Project
We propose Periscope3D – a robotic system that replaces traditional 2D camera feeds with real-time 3D spatial reconstruction to enable more immersive and spatially-aware remote collaboration. Current remote physical collaboration systems typically involve a remote expert (helper) guiding a local worker through physical tasks via a robot-mounted 2D camera, but suffer from occlusion issues, viewpoint constraints, and split attention between task execution and camera management.
Our proposed system integrates multiple 3D sensing modalities to create a real-time 3D representation of the workspace that can be explored intuitively by remote collaborators through VR/AR interfaces, while maintaining the proven shared control mechanisms. This approach aims to overcome fundamental limitations of 2D-based robotic collaboration by providing complete spatial awareness, eliminating viewpoint constraints, and enabling independent exploration of the workspace without disrupting the local worker's activities.
Project Duration
- 10 - 12 weeks
Required Skills
- Programming skills in C++ and/or Python under Linux
Preferred experience
- Experience with ROS (Robot Operating System)
- Previous work with computer vision libraries and/or depth sensing technologies
Seeing the unseeable: super-resolution of echocardiograms
Project code: ABI029
Supervisor(s):
Dr Gonzalo Maso Talou
Prof. Martyn Nash
Discipline:
Auckland Bioengineering Institute
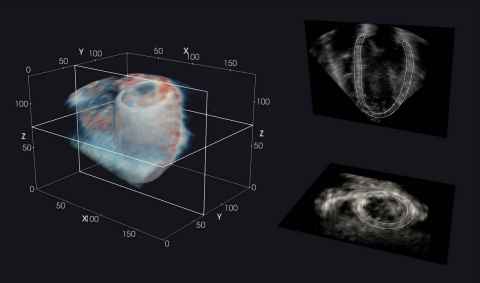
Project
Cardiac ultrasound (or “echocardiography”) is the mainstay for non-invasive clinical assessment of the heart. However, compared to other imaging techniques, echocardiograms are typically of low contrast-to-noise ratio and spatial resolution.
The role
The aim of this project is to leverage deep learning models which have been successful in other imaging domains, such as cinematography, to “super-resolve” echocardiograms. This may help to provide a clearer visualisation of the heart in health and disease.
You will join a multi-disciplinary team of bioengineers, computer scientists, clinicians, and imaging specialists.
Ideal student
This project will suit students with an interest in artificial intelligence, medical image analysis, and translational research. Prior medical imaging experience is not required.
Investigating how and where the breast moves using AI
Project code: ABI030
Supervisor(s):
Poul Nielsen
Martyn Nash
Thiranja Prasad Babarenda Gamage
Supported by ABI breast biomechanics research group
Discipline:
Auckland Bioengineering Institute
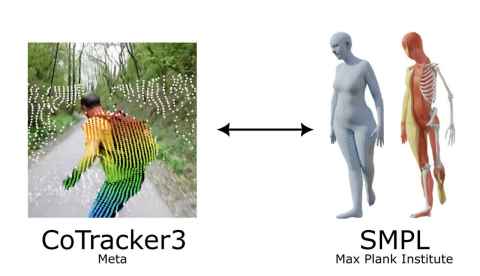
Project
Breast cancer affects 1 in 9 women in Aotearoa New Zealand. A key challenge for clinicians is determining where tumours move as the breast deforms due to changes in patient positioning during treatment procedures, such as surgery or radiotherapy. Tracking arm movements and the deformations of the patient’s skin surface provides useful information to biomechanical models that are being developed to predict the position of tumours within the breast during repositioning.
The role
This project aims to combine cutting edge machine learning-based motion tracking algorithms with whole body modelling tools to accurately track the position of the shoulder and breast during clinical procedures. You will join a multi-disciplinary team of bioengineers, clinicians, computer vision experts and instrumentation specialists.
Ideal student
This project will suit students with an interest in computer vision, machine learning and computer programming.
Too thin to matter? Deep learning for 3D modelling of the right ventricle
Project code: ABI031
Supervisor(s):
Joshua Dillon
Dr Debbie Zhao
Discipline:
Auckland Bioengineering Institute
Project
Cardiac magnetic resonance imaging is the gold-standard technique for anatomical imaging of the heart. While much attention has been directed towards the left ventricle, the right ventricle (RV) is comparatively understudied. The mass of the RV has recently emerged as an important index of heart remodelling. However, RV mass is controversial and challenging to calculate owing to the thinness and complexity of the RV wall.
The role
The aim of this project is to refine a deep learning framework to accurately estimate RV mass from CMR, and explore its diagnostic and/or prognostic value in a real-world clinical cohort. As part of a NZ Government funded programme on clinical translation of heart modelling, outputs of this project will contribute to more efficient and effective strategies for the diagnosis and monitoring of heart disease.
You will join a multi-disciplinary team of bioengineers, clinicians, and imaging specialists.
Ideal student
This project will suit medical or engineering students with an interest in artificial intelligence, medical image analysis, and translational research. Prior medical imaging experience is not required.
Going with the flow: estimating heart pressures non-invasively
Project code: ABI032
Supervisor(s):
Mathilde Verlyck
Debbie Zhao
Discipline:
Auckland Bioengineering Institute
Project
Cardiac magnetic resonance imaging (MRI) provides detailed, non-invasive views of the heart, and is considered the gold standard for assessing cardiac size and function. However, routine cine MRI does not capture blood flow velocities, which may help to estimate heart pressures without invasive procedures. This is particularly important for the management of heart failure, a condition affecting about 80,000 New Zealanders.
The role
This project aims to leverage computer vision approaches to assess blood flow velocities from MRI, and to create a modelling approach for non-invasive estimation of left ventricular pressures.
You will join a multi-disciplinary team of bioengineers, clinicians, and imaging specialists.
Ideal student
This project will suit students with an interest in computer vision, medical image analysis, and translational research. Prior medical imaging experience is not required.