Biological Sciences
Applications for 2025-2026 open on 1 July 2025
Evaluating the magnet effect in Aotearoa’s ecosystems: are exotic plants good or bad for pollination services?
Project code: SCI042
Supervisor(s):
Sandra Anderson
Discipline(s):
School of Environment
School of Biological Sciences
Project
The magnet effect refers to attractive flowering plants drawing in pollinators, which can benefit or distract from neighbouring species.
The role
This project will evaluate the potential for magnet effects in Aotearoa-New Zealand by identifying susceptible native plant species based on their phenology, distribution, and pollinators. We will review global literature to understand the conditions under which magnet effects occur. These insights will help us assess their relevance in New Zealand ecosystems and develop methods for empirical field evaluation.
Exploring the role of traditional harvesting practices in sustaining in-stream macrophyte ecosystems
Project code: SCI061
Supervisor(s):
Cate Macinnis-Ng (SBS)
Dan Hikuroa (MAORI STUDIES)
Sarah Rewi (SBS)
Discipline(s):
Ngā Motu Whakahī
Biological Sciences
Project
This is a Ngā Motu Whakahī scholarship specifically targeted toward students who whakapapa Māori or are of Pacific heritage.
This project will investigate freshwater ecosystem management drawing from mātauranga and science.
The role
Research will be fieldwork-based, looking into population dynamics of watercress in rural streams. It will examine the impact of different harvesting techniques as strategies to promote growth and removal of pollutants.
Ideal student
Ideal for students that have an interest in ecology and the interface between mātauranga and science.
Requirement
Whakapapa Māori is required.
Variation in myrtle rust susceptibility of pōhutukawa in Auckland city
Project code: SCI097
Supervisor(s):
Discipline(s):
School of Biological Sciences

Project
Myrtle rust (Austropuccinia psidii) is a virulent plant pathogen of the family Myrtaceae, with the rust now having spread globally and infecting >500 plant species. It arrived in New Zealand in 2017 and is still spreading. Because it is a recent arrival, our understanding of host susceptibility and disease progression is also still developing, both within and amongst species. For pōhutukawa within Auckland, infections of individuals seem to vary by infection date, location, plant size, proximity to roads, and whether the plant was planted or wild grown. Sometimes infected plants and healthy plants occur directly next to each other.
The role
This proposed project would build on previous surveys of pōhutukawa within Auckland to score the infection status of trees and a range of potential factors (e.g., size, proximity to roads, and likely origin) that influence susceptibility to infection. These data would then be analysed for infection patterns both within and between years. The goal is to determine factors (host, environment) that determine levels of myrtle rust infection to better understand the infection process and inform management response.
Impact of sleep disturbance on bird behaviour
Project code: SCI098
Supervisor(s):
Discipline(s):
School of Biological Sciences
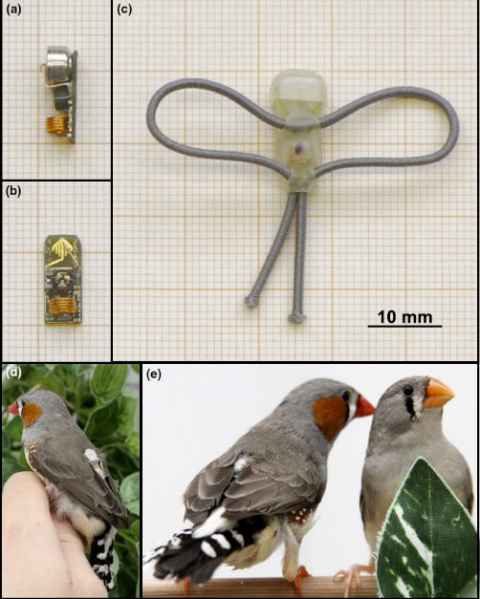
Project
Diurnal birds sleeping in the wild are constantly being disrupted by stimuli such as artificial light at night and anthropogenic noise. As humans, a bad night of sleep can negatively affect our function, but we often don’t think about what it may be like for animals.
The role
The student will analyse behaviour of sleep disturbed birds. the student will help with husbandry and daily care of animals; therefore, they must be able to come to campus during the week.
Stress impacts of climate change on NZ seabirds
Project code: SCI100
Supervisor(s):
Ms Inka Pleiss (PhD candidate)
Discipline(s):
School of Biological Sciences

Project
Climate change causes stress levels in seabirds to increase. One keyway to measure this is to measure blood cell ratios (Heterophil: Lymphocyte ratio) and AI methods are being developed to achieve this.
The role
This project requires a student to train and test an AI model to count blood cell smears on slides (H:L slides) taken from tītī across Aotearoa and life stages
Required skills
Ability to use a microscope, diligence and thoroughness. There will be opportunity to assist with field studies, but this is primarily a lab based project.
Nitrogen stockpiling in cyanobacteria
Project code: SCI101
Supervisor(s):
Discipline(s):
School of Biological Sciences
Project
Some cyanobacteria can store nitrogen in granules (cyanophycin) inside their cells. Our lab works with a cyanobacterial genus that can do this. However, some members are predicted based on genetics to stockpile more nitrogen.
The role
In this project a student will undertake laboratory based cultivation experiments to help confirm if this is true, and the impact on survive under nutrient deprivation.
Required skills
A background in microbiology with good laboratory skills is essential, alongside an interest in microbial cultivation and cell imaging (microscopy).
Some like it salty: evolution of salt tolerance in estuarine bacteria
Project code: SCI102
Supervisor(s):
Discipline(s):
School of Biological Sciences
Project
This project will investigate the metabolic responses of estuarine bacterial isolates grown at different salinities. The isolates are conspecific (same species). Initial growth experiments suggest they have evolved different salt tolerances.
The role
The goal of this project is to confirm or disprove the evolution distinct salt tolerances within the species. This project will involve analysis of a genomic and transcriptomic dataset.
Requirements
A background in microbiology, good or promising computational skills, and an interest in bioinformatics and genomics are essential.
Peptide affinity selection mass spectrometry for rapid peptide drug discovery
Project code: SCI103
Supervisor(s):
Assoc Prof Paul Harris
Dr Daniel Conole
Dr Renata Kowalczyk
Discipline(s):
School of Biological Sciences
Project
Peptide affinity selection mass spectrometry (Pep AS-MS) is an exciting new high throughput drug screening technique that utilises high resolution nano-liquid chromatography mass spectrometry (nano LC-MS/MS) and bioinformatics.
The role
In this project, the student will design and perform on-resin combinatorial peptide chemistry routes to design bespoke and complex peptide libraries. This project could lead into further post-graduate study where the synthesised peptide library is screened against an important drug target in inflammation, and hit peptide binders synthesised individually, and their binding validated.
Ideal student
This project is ideal for someone with strong interests in either peptide chemistry, chemical biology or bottom-up proteomics/mass spectrometry.
Smart Cleavable Linkers for Enhanced Precision and Safety of Antibody–Drug Conjugates in Cancer Therapy
Project code: SCI104
Supervisor(s):
Discipline(s):
School of Biological Sciences

Project
This project aims to design, synthesise, and evaluate smart cleavable linkers for antibody–drug conjugates (ADCs), one of the fastest-growing classes of targeted cancer therapies. Traditional linkers often trigger premature payload release in healthy tissues, leading to off-target toxicity and limiting clinical efficacy.
The role
By incorporating stimuli-responsive elements that are activated specifically within the tumour microenvironment, this research seeks to improve release precision, expand the therapeutic window, and reduce systemic toxicity—addressing a key challenge in the safe and effective application of ADCs in cancer treatment.
Multifunctional Peptides for Precision Cancer Theranostics
Project code: SCI105
Supervisor(s):
Discipline(s):
School of Biological Sciences
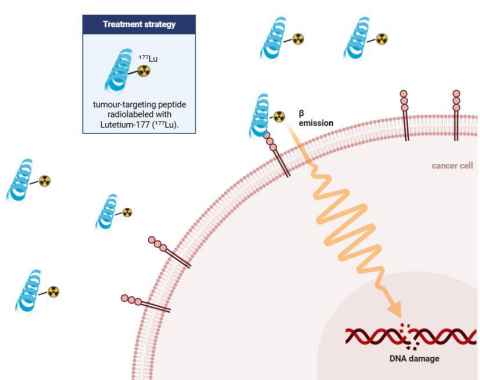
Project
This project aims to develop peptide-based theranostic agents that combine targeted cancer therapy with diagnostic imaging. Through rational peptide design and chemical synthesis, we will create multifunctional peptides that specifically bind tumour biomarkers while carrying both therapeutic payloads and imaging reporters. The peptides will be optimised for high affinity, stability, and selective tumour accumulation. This integrated approach seeks to enable real-time monitoring of drug delivery and treatment response, advancing personalised and precision cancer care.
What does the choanoflagellate genome encode?
Project code: SCI106
Supervisor(s):
Discipline(s):
School of Biological Sciences
Project
Choanoflagellates are often overlooked aquatic hunters of bacteria, and the closest living relatives of animals. For the past six years, students in our final year undergraduate protein science paper, BIOSCI350, have been sequencing genes, and producing proteins from the model choanoflagellate Salpingoeca rosetta. We have identified a number of interesting choanoflagellate proteins which we can make in large amount.
The role
In this summer project you’ll attempt to isolate and structurally characterize some of these proteins.
Investigating metabolic enzymes in cancer: recombinant expression and small molecule screening
Project code: SCI107
Supervisor(s):
Ben Krinkel (PhD candidate)
Discipline(s):
School of Biological Sciences
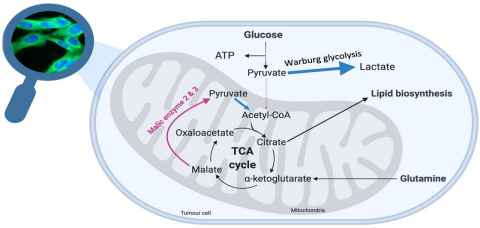
Project
Cancer cells exhibit remarkable adaptability, evading the usual cellular checkpoints to fuel their relentless proliferation. Our research focuses on two metabolic enzymes, malic enzymes and Aspartate aminotransferase, critical players in cancers altered metabolism. By investigating the structural nuances of malic enzyme isoforms, we aim to uncover vulnerabilities that can be exploited for targeted anticancer therapies.
The role
This project will encompass the expression and purification of human malic enzyme isoforms utilizing an existing bacterial expression system. Prior structural elucidation have facilitated the acquisition of small molecules. These molecules are slated for screening against the three human malic enzyme isoforms to investigate the complex binding interactions with small molecule inhibitors. Contingent upon progress, protein crystallography may commence for any molecules that demonstrate binding affinity.
Ideal student
Skills in protein expression, however, all applicants will be considered!
Searching for new antibiotics
Project code: SCI108
Supervisor(s):
Discipline(s):
School of Biological Sciences
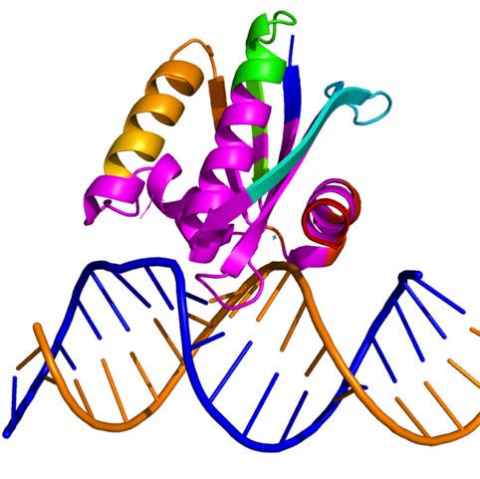
Project
Resistance to antibiotics is a growing global problem, which is estimated to place 10 million lives per year at risk by 2050 without action. This project will focus on understanding an enzyme (RNase HI) that is a target for new antibiotics against Neisseria gonorrhoeae.
Ideal student
This project is best suited to someone with a strong interest in protein structure and function and/or microbiology.
The role of RhsA in bacterial competition
Project code: SCI109
Supervisor(s):
Discipline(s):
School of Biological Sciences
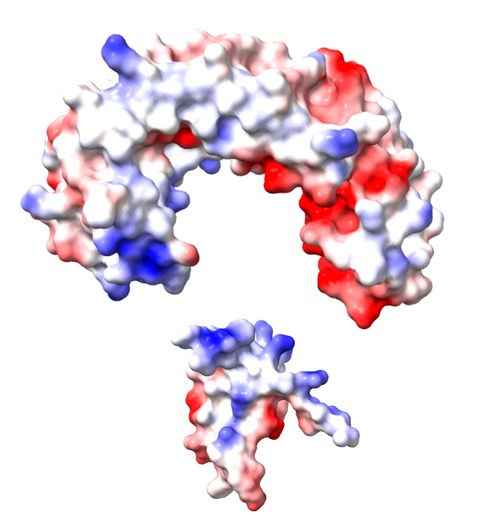
Project
RhsA is a conserved bacterial protein involved in bacterial competition by delivering a toxic cargo protein to adjacent cells, probably via the Type VI secretion system. What we don’t know is why the protein is cytotoxic.
The role
This project will use mutant proteins to test ideas about the function of RhsA, based on our recently determined structure of the protein.
Ideal student
This project is best suited to someone with a strong interest in protein structure and function.
Plant ecophysiology in a changing climate
Project code: SCI110
Supervisor(s):
Discipline(s):
School of Biological Sciences

Project
Plants are highly responsive to environmental conditions, including climate change but our knowledge of impacts of stressors like drought and heat on the flora of Aotearoa is still emerging.
The role
This summer student will assist two postgraduate students working in the field and glasshouse assessing the vulnerability of native plants to water and warming stress.
Requirement
A strong interest in plant biology is required. Training will be provided.
Protein structure phylogenetics methods
Project code: SCI111
Supervisor(s):
Discipline(s):
School of Biological Sciences
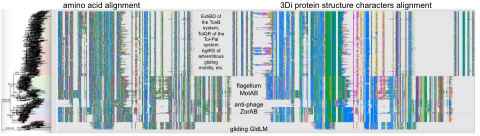
Project
The project involves work on some aspect of our new methods for doing phylogenies of remotely-related sequences using protein structure. For example: comparison of available methods for alignment.
Desired skills
Prior experience with R, using your computer’s command line, basic bioinformatics or phylogenetics.
Phylogeny of the bacterial flagellum and related systems
Project code: SCI112
Supervisor(s):
Discipline(s):
School of Biological Sciences
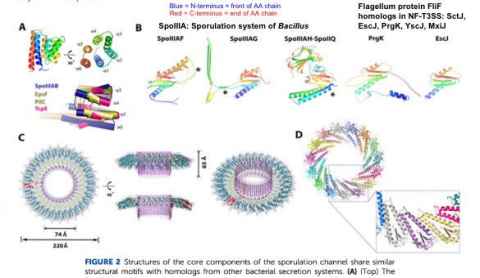
Project
The flagellum is a complex adaptation made of 40-50 different proteins, and explaining its evolution is a famous puzzle. Our lab has been working through these proteins to use new methods and new homologous outgroups to learn more about its history.
Desired skills
Prior experience with R, using your computer’s command line, basic bioinformatics or phylogenetics.
Exploring clinical trials in nutrition and metabolism
Project code: SCI113
Supervisor(s):
Assoc Prof Jennifer Miles-Chan
Discipline(s):
School of Biological Sciences

Project
Eager to dive into the exciting world of human clinical trials? Join the University of Auckland Human Nutrition Unit for an immersive summer studentship!
The role
You'll gain unique insight into translating nutritional science into real-world health applications. Working within our close-knit team in Mt Eden, and depending on projects running at the time, you could be involved in participant interaction, project planning, study design, or data wrangling
Required skills
Great communication skills, attention to detail, and plenty of enthusiasm are key!
Daily Temperature and Carbon dioxide rhythms Patterns in Honey Bee Colonies
Project code: SCI114
Supervisor(s):
Dr Rose McGruddy
Dr James Cheeseman
Discipline(s):
School of Biological Sciences
Project
Robust circadian rhythms are essential to the health of all living organisms, and the breakdown of these rhythms can predict disease or even death. Honey bees are no exception. Their entire colony structure is governed by the circadian clock.
The role
This project focuses on understanding daily rhythms of temperature and carbon dioxide (CO₂) inside the hive. You will collect and analyse high-resolution data to establish:
(1) Baseline rhythms in temperature/ CO₂
(2) How rhythms change following controlled disturbances and
(3) How changes may be used to predict upcoming colony events.
Validating the electrostatic signatures of waggle dances in honey bee hives
Project code: SCI115
Supervisor(s):
Dr Rose McGruddy
Dr James Cheeseman
Discipline(s):
School of Biological Sciences
Project
Our understanding of how the honey bee clock functions remains limited – largely because direct observation requires opening the hive, which disrupts behaviour.
We have developed a novel high-resolution electrostatic field (ESF) sensor system (BeeSpy) that allows us to non-invasively monitor bee behaviour inside closed hives.
The role
This project will focus on validating ESF signals of waggle dancing bees against video footage recorded from observation hives. You will collect and analyse concurrent video and ESF recordings of waggle dances, and use AI-based systems to identify characteristic ESF signatures. These signatures will then be used to identify waggle dances in undisturbed hives.
How does genetic variation in clock genes link to local environmental conditions?
Project code: SCI116
Supervisor(s):
Discipline(s):
School of Biological Sciences
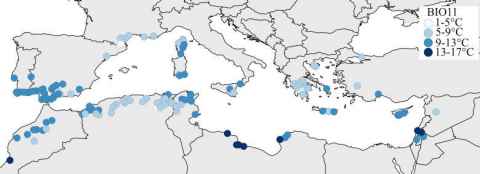
Project
Legumes rank just behind cereals as one of the world’s most important crop groups, including vital species like lentils, peas, and lucerne. To thrive and adapt, these plants depend on their internal timekeeper—the circadian clock—which coordinates daily rhythms in essential processes like growth and photosynthesis. By understanding how this clock operates in diverse environments, we can develop legume crops that better withstand climate change.
The role
This project explores how variations in the DNA sequences of circadian clock genes in the model legume Medicago truncatula connect to the temperature conditions where different accessions originally grew. By analysing public climate data alongside genomic information from diverse natural accessions, the project aims to uncover clear links between temperature and specific gene variants.
To confirm these findings, the project includes a hands-on physiological study that measures clock function at a controlled temperature in selected accessions with contrasting gene types.
Skills gained
- Develop skills in analysing genomic and climate data using R.
- Carry out and interpret experiments on plant traits controlled by the circadian clock.
Ideal student
A student with a strong interest in genomics and plant physiology. BIOSCI 202, 205, 351, and 326 provide a good background.
Analysing changes in pressure-volume loops during lung injury
Project code: SCI117
Supervisor(s):
Dr Victor Zimbron
Discipline(s):
School of Biological Sciences
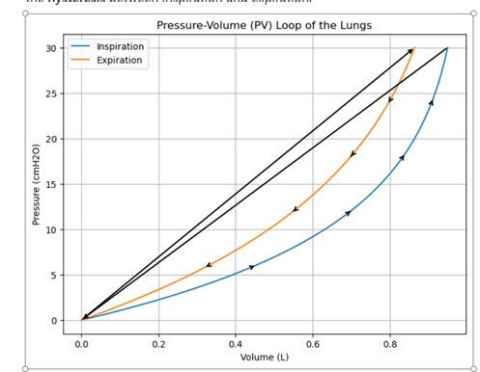
Project
During mechanical ventilation, Pressure-volume loops (PV-loops) are helpful to understand how lungs respond to injury. This project explores different ways to compare and visualize repeated PV-loop data collected from preclinical models.
The role
You will help develop new analytical methods to track changes, spot trends, and highlight meaningful patterns in PV-loops.
Ideal student
Ideal for students of biology with some basic lung anatomy understanding as well as comfort working with computers, coding and keenness to gain hands-on experience in biomedical data analysis and visualization.
Aims
The project aims to create a more efficient way to use this data generated from a ventilator by:
A) creating some automatic analysis of curves so it is not manual
and/or
B) developing a visualisation and analysis methodology for a 3D PV-loop (pressure-volume-time)
Proof of concept for a novel mechanical ventilator
Project code: SCI118
Supervisor(s):
Dr Victor Zimbron
Discipline(s):
School of Biological Sciences
Project
This hands-on project involves assisting in building and testing a new type of mechanical ventilator. You’ll work closely with a small team, learning about lung biomechanics and bioengineering while solving real-world biomedical problems.
Ideal student
This opportunity is ideal if you're creative, enjoy troubleshooting, and want direct experience turning a novel medical concept into reality.
Requirements
The ideal background requirements are an interest in basic biology (especially lungs) and device engineering. No specific formal university engineering course experience is expected, however self-taught basic technical skills are a definite advantage. An interest in making devices and experimenting with them is expected.
The coastal meroplankton of Rangitāhua (the Kermadecs)
Project code: SCI119
Supervisor(s):
Discipline(s):
School of Biological Sciences
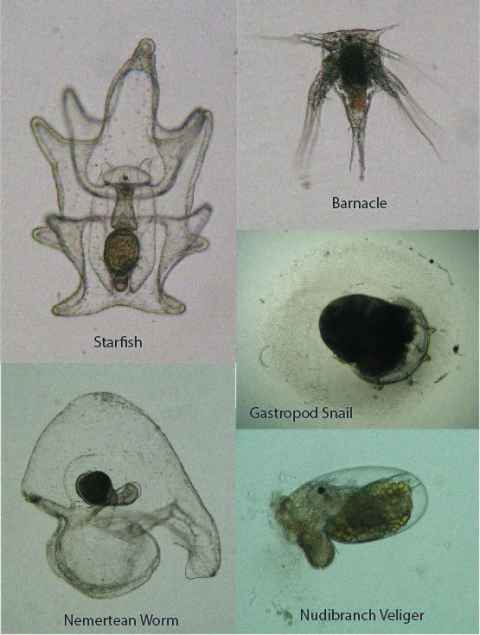
Project
Plankton samples that have been at the Kermadecs (MBIE project: Te Mana o Rangitāhua: Ngati Kuri and Auckland Museum) will be examined using a microscope, with a focus on marine invertebrate and fish larvae.
Ideal student
An understanding of marine invertebrate diversity would be a distinct advantage (e.g., completion of BIOSCI 208 or similar), but on-the-job training will be provided. Ideally suited for a student with interests in biodiversity, identification of small organisms, and in gaining skills in scientific photography and illustration.
Urban birds: are they spreading weeds?
Project code: SCI120
Supervisor(s):
Discipline(s):
School of Biological Sciences

Project
This project continues some of our research on urban bird communities and their interaction with exotic plants. The project may include bird observations, some vegetation surveys and some GIS mapping. You will work as part of a team with our postgraduate students studying urban ecosystems.
Requirements
Must have driver’s licence (preferably full) and access to a car.
Exploring Protein–Chemotherapeutic Agent Interactions
Project code: SCI121
Supervisor(s):
Discipline(s):
School of Biological Sciences
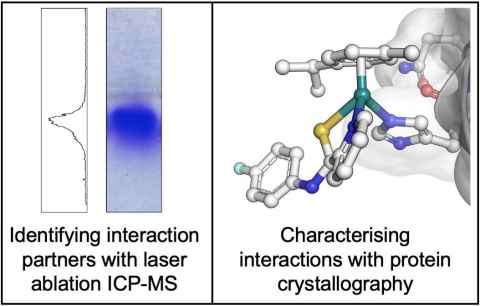
Project
A novel antiproliferative agent plecstatin-1 has been shown to selectively target the protein plectin a known regulator of malignant phenotypes. Plectin is a crucial cytoskeletal component, which acts as a cytolinker.
The role
My research project explores the interaction between plecstatin-1 and plectin to understand how selectivity is derived and to understand the role that plectin has within the cytoskeleton. The bioanalytical methodologies surface plasmon resonance and a variety of mass spectrometry techniques (example shown on the left side of the image) will be used to identify the binding domains and regions involved in the plectin–plecstatin-1 interaction.
Through protein crystallography (example shown on the right side of the image) and specific binding assays, detailed binding mechanisms and affinities governing the plectin–plecstatin-1 interaction and how this is interrupts the cytoskeleton will be uncovered.
The overarching goal is to elucidate how plecstatin-1 interacts with plectin, which in turn interrupts the cytoskeleton and thereby cancer cell behaviour, providing crucial insights into the molecular mechanisms driving cancer progression. Ultimately, this deeper understanding holds potential for the development of targeted cancer therapeutics, addressing an unmet need in clinical practice.
Ideal student
No specific skills are required but this project would best suit to someone with an interest in proteins and how they interact with therapeutic agents.
Identifying novel therapeutic targets from the world's deadliest pathogen
Project code: SCI122
Supervisor(s):
Assoc Prof Ghader Bashiri
Discipline(s):
School of Biological Sciences
Project
Tuberculosis remains the leading cause of death worldwide from a single infectious agent. The bacterium responsible for tuberculosis has evolved to scavenge carbon sources such as fatty acids from human host cells to sustain chronic infection.
The role
This research aims to investigate how the bacterium regulates its metabolism, identifying novel targets for therapeutic intervention.
Ideal student
Students with a background in protein science will be suitable for this position.
Skills gained
During this project, students will gain hands-on experience in protein expression, purification, and biochemical assays.
This project is funded by the Marsden Fund to support applicants who whakapapa Māori.
Indexing the Overlooked: Addressing Invertebrate Omissions in Genomic Reference Databases
Project code: SCI123
Supervisor(s):
Dr Neil Birrell
Prof Jacqueline Beggs
Discipline(s):
School of Biological Sciences

Project
DNADRV uses traces of insect DNA from car numberplates to infer insect diversity across Aotearoa. However, identifying many insects is stymied by extremely poor reference databases, especially outside of Northern America and Wealthier European countries.
The role
This summer project involves following a protocol to sequence insect mitogenomes. Researching and selecting under-represented taxa comprises the bulk of the project – including meeting with taxonomic experts. Mitogenome assembly and annotation is optional.
Ideal student
No skills or pre-requisites, just interest in learning molecular approaches to ecosystem conservation.
Biodiversity of insects in a taxonomic collection at Landcare Research
Project code: SCI124
Supervisor(s):
Discipline(s):
School of Biological Sciences
Project
Two students
Work with entomologists and taxonomists on insect projects from New Zealand and the Pacific.
Learn research and technical skills that will allow you to work in a taxonomic collection or natural science museum, including:
1) fieldwork and sorting field samples
2) curation and identification of insect groups
3) imaging specimens for machine learning / AI methods
4) databasing and geo-referencing specimens for biodiversity and biosecurity research projects
Requirements
Ability to work in a group or independently; position requires dexterity, care and attention to detail, as well as data entry. Applicants should have an interest in Entomology, Taxonomy, Natural History
All things Fungi: learning about mycology and plant pathology in a nationally significant fungarium
Project code: SCI125
Supervisor(s):
Discipline(s):
School of Biological Sciences

Project
Two students
The Landcare Research fungal systematics group maintains the largest collections of both dried and living fungi in New Zealand, and delivers data associated with the collections through the BiotaNZ website.
The role
The two successful applicants will provide assistance adding data to the fungal database, and maintaining, tracking, and storing fungal specimens and cultures and associated DNA sequence data.
Skills gained
You will gain an understanding of the day to day running of a mycological research laboratory and key techniques you will learn include handling fungarium specimens, data capture and management, and microscopy.
In addition, you will be expected to assist with ongoing projects in the mycology labs at Manaaki Whenua-Landcare Research, Tamaki. These could potentially involve culturing and identification of fungi from field samples, analysis of DNA data, etc.
Requirements
Attention to detail, ability to keyboard accurately, a general interest in fungi, bacteriology or plant pathology.
Impact of sleep disturbance on bird song
Project code: SCI165
Supervisor(s):
Discipline(s):
School of Biological Sciences
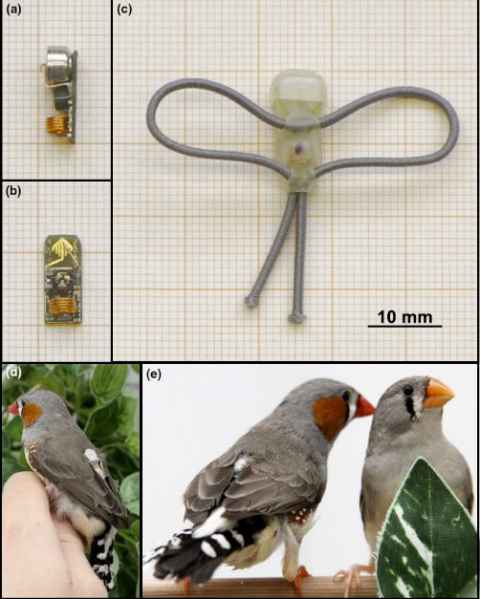
Project
Diurnal birds sleeping in the wild are constantly being disrupted by stimuli such as artificial light at night and anthropogenic noise. As humans, a bad night of sleep can negatively affect our function, but we often don’t think about what it may be like for animals.
The role
The student will analyse song of sleep disturbed birds. Student will help with husbandry and daily care of animals; therefore, they must be able to come to campus during the week.