Enabling development and clinical translation of virtual human twins
On this page:
DigitalTWINS platform
As part of the 12 Labours project, an open-source collaborative platform called DigitalTWINS (Digital Translational Workflows for Integrating Systems) was developed. The platform was designed to provide shared infrastructure and services to assemble computational physiology workflows for creating personalised digital twins. It also aimed to support researchers conducting clinical studies to evaluate the performance and clinical translation of these workflows.
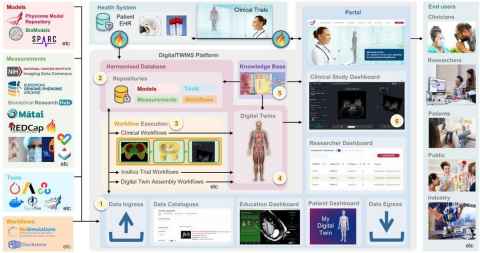
Infrastructure and services developed for the platform included:
- Uploading of data such as health research records, clinical measurements, computational models, tools, and existing workflows.
- Secure, access-controlled, and harmonised data storage.
- Assembly and execution of clinical workflows to personalise computational models (digital twins) for specific clinical applications.
- A web portal that provided secure access for multiple user groups, including:
- Data catalogues (“yellow pages”) to help users locate datasets for reuse.
- Researcher dashboards to allow rapid testing of workflows on clinical measurements.
- Clinical research study dashboards where clinicians and researchers could jointly assess workflow efficiency.
- Patient dashboards where individuals could view their own digital twins generated from their data.
- Education dashboards to visualise complex physiological concepts for learning and public engagement. - Connection to health systems to streamline clinical studies and accelerate translation of digital twin workflows.
The adoption and support of this platform had the potential to foster an ecosystem for reproducible research outcomes and to provide a foundation for integrating research efforts toward the clinical application of digital twins.
A prototype was implemented on the ARDC Nectar Cloud, supported by the University of Auckland Centre for eResearch and the New Zealand eScience Infrastructure (NeSI). It was tested with the four clinical workflows developed during the 12 Labours project, with a production version planned for wider ABI adoption. The platform was also envisioned as a potential national initiative, for example, in support of the MedtechIQ programme.
DigitalTWINS repository
Developing technologies such as digital twins to improve health, well-being, and quality of life required addressing the grand challenge of understanding how organs interact as part of whole-body systems. Achieving this demanded detailed and standardised datasets linking specialised research measurements, biobank samples, and national health registers.
The 12 Labours DigitalTWINS Repository was established as a pioneering initiative to create such a data resource—enabling health information from participants across multiple research studies to be contributed, linked, and reused with informed consent. This resource was designed to deepen understanding of human physiology and to underpin technological innovation, supporting digital twins of individuals, hospitals, and populations for improved health, social, and economic outcomes.
Key goals and components included:
- Establishing consistent procedures and infrastructure underpinned by data sovereignty principles for contributing and accessing data.
- Providing standardised protocols and systems for linking datasets across studies.
- Enabling the secondary use of previously contributed data for new research, once appropriate approvals were obtained.
- Supporting future participant contact for ongoing data collection and linkage.
- Implementing data management consistent with FAIR principles (Findable, Accessible, Interoperable, Reusable).
The Repository structure included:
- A Data Contribution and Access Committee (DCAC) to assess data contribution and access requests.
- A Data Management Plan outlining secure, ethical, and responsible data handling practices aligned with data sovereignty and consent principles.
- A four-tier Data Platform:
1. Restricted Access, Identifiable Data Tier – storing participant recruitment and consent data.
2. Restricted Access, De-identified and Linked Data Tier – storing linked datasets under a de-identification protocol.
3. Restricted Access, De-identified and Unlinked Data Tier – providing approved data subsets for research teams.
4. Public Access Data Tier – containing non-sensitive metadata and any openly available datasets.
- A Governance Board, guided by Terms of Reference, to oversee the Repository.
The Repository’s long-term vision was to ensure that participants’ data would continue to positively impact the well-being of current and future generations (mokopuna).
A New Zealand Health and Disability Ethics Committee (HDEC) application was prepared in consultation with community groups and partner organisations and was undergoing internal University of Auckland review prior to submission for ethics approval.