Animal and Microbiome
Integrating experiments, computational modelling and instrumentation to improve understanding of host-microbe interactions in humans and animals.
Systems models of the rumen microbiome and metabolome
In the rumen, metabolic cascades are carried out by the microbial community in a complex and coordinated manner, whereby successive food webs of cross feeding exist between different rumen microorganisms. These cascades supply basic metabolites to the hosting animal. Indeed, microbial production of both short- and long-chain fatty acids serves up to 70% of the animal’s energy needs.
Microbial ecology of the rumen has long been the focus of research. The Hungate1000 project assembled virtually all the bacterial and archaeal species that have been cultivated from the rumens of a diverse group of animals. However, such methods provide little mechanistic insight into how the dynamic couplings between individual microorganisms give rise to their dynamic activities.
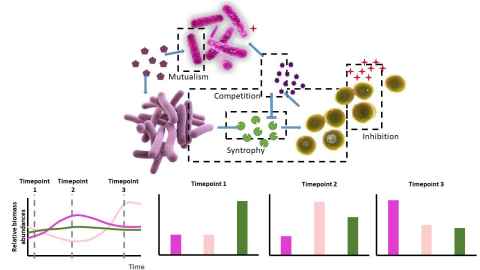
A key question is therefore to determine what can and cannot be inferred from only metagenomic data and knowing this, what additional experimental measurements and computational methods are required to get a more complete understanding of a microbial ecosystem. This challenge is largely solved for single species where systems biology has delivered many methods for inference, data integration, and predictive modelling. These methods have become useful tools in more applied fields such as synthetic biology and metabolic engineering. Our work focuses on developing inference methods to determine microbial ecosystem and the causal interactions among the various microbes and substrates.
Precision livestock farming
Animal husbandry revolves around the relationship between farmers and their animals. Farmers are required to ensure the very best of care for their animals, whilst trying to optimise their production cycles, lactational performances, and muscle development in a cost-effective way.
Animal wellbeing is multifactorial, and achieving the best solutions require inputs from animal scientists, veterinarians and agricultural engineers, working in partnership with equipment companies. Precision livestock farming consists of measuring variables, modelling the data to select information, and then using these models in real-time for monitoring and controlling the animals. The main variables that might be monitored are the use of key livestock resources (feed, water, land surface), performances (milk yield, weight), health (for instance coughing, body temperature, lameness), behaviour (activity, rumination, fighting) and environment (temperature, humidity, air speed, toxic gases).
Critical to integrating and analysing this data is the underlying model of the cow and the richness of its features to assimilate the inputs of various sensors, enable causal investigations and capability to predict future states for known interventions. This suggests that the development of a Reference Cow model is necessary. We are developing a comprehensive computer model that captures the key physiological mechanisms of the cow; that will enable integration and analysis of the sensorial data.
Biogeography of the gut/skin microbiome
The gut and skin contain a number of distinct habitats that select and host a spatially heterogeneous community of microbes.
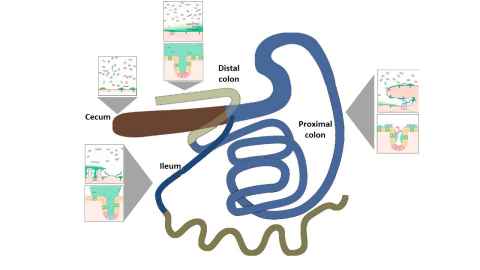
Recent studies have shown spatial reprogramming of the microbiota to be a functionally relevant feature in chronic inflammatory diseases. Determining the key factors that influence this spatial organization is essential for addressing several chronic diseases and improving general well-being. Multitude of factors such as chemical gradients, nutrient availability, biomechanical processes, and immune effectors have been hypothesized to drive heterogeneity. Experimental devices mimicking these environments provide highly valuable information on the function of the microbiota and their spatial structure. While reductionist approaches on gnotobiotic rodents are highly valuable tools, they only partially mimic the host response or the ecosystem functions. This makes it difficult to evaluate the relative importance of the factors that determine biogeography. Our group is developing physiologically realistic mathematical models to complement experiments and gain insight on the main factors.
Rumen Bioreactor
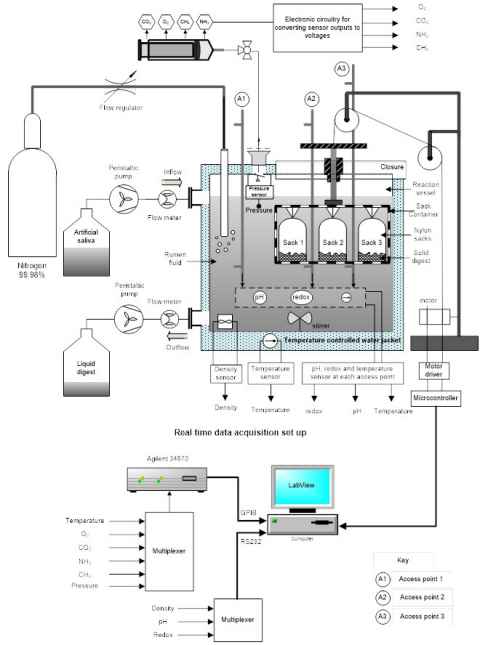
One of the challenges in systems modelling of the microbiome is the high level of variability in data acquired from field samples, which results from differences between individual animals, their environment and sampling conditions. We are developing bioreactor models of the rumen to create model systems in which the inputs and outputs can be precisely controlled and monitored. This will reduce variability from confounding factors and make it possible to carry out more detailed measurements to produce richer datasets to support mechanistic modelling. The bioreactor will provide an anaerobic environment that mimics the rumen to support the growth of rumen microbes. Automated flow control, in-built sensors and access points will enable precise control of experimental conditions and the sampling of contents to characterise microbial community and chemical composition.
Modelling physiome-microbiome interactions
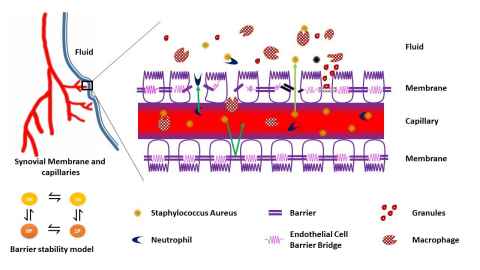
The healthy human body is teeming with microorganisms, many of which play essential roles in our metabolism, our immune response, and general wellbeing. Under adverse conditions, these very organisms turn pathogenic and compromise host health. Understanding the host-microbiome interactions may also help to explain why some of us are prone to certain illnesses and therefore vital for advancing personalized medicine. We are developing mathematical models and frameworks for investigating the interactions between human and microbial systems. Systems of interest include Periprosthetic Joint implants, Atopic dermatitis and microbial endocrinology.
Opportunities
Funded PhD positions, internship and summer scholarship opportunities are available. Expressions of interest should be sent to:
Dr. Vinod Suresh
Dr. Jagir Hussan
-
Animal and microbiome opportunities poster. Size: 2.0 MB.
Document Description: Join our world-leading team driving the latest advances in animal and microbiome research
Members
Primary contacts
Academics
Prof. Peter Hunter
Students
Chernet Woyimo Woju
Collaborators
Prof. Ian Hunter (BioInstrumentation Lab at MIT)
Dr Mark Oliver
Prof. Jillian Cornish
Funding
- Callaghan Innovation
- Fonterra Cooperative Group